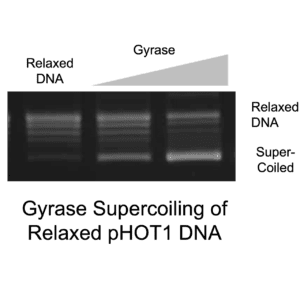
Relaxed pHOT-1 is a DNA Plasmid suited to assaying the supercoiling activity of DNA gyrase. It is also useful for assaying alterations in DNA linking number for assessing intercalation. The plasmid is prepared using high purity topo I relaxation reactions, followed by inactivation and re-purification of the relaxed product. TopoGEN staff then verify the relaxation status of each lot and validates that the substrate can be supercoiled by E. coli DNA gyrase and S. aureus DNA gyrase.
The plasmid pHOT-1 contains the high affinity topo I cleavage site (the hexadecameric sequence described by Westergaard and co-workers, see Bonven et al., 1985) that is derived from the tetrahymena ribosomal gene repeat. This construct is useful as a topo I cleavage substrate since the enzyme efficiently cleaves at the hexadecameric site in the absence of camptothecin. This allows the investigator to rigorously evaluate new potential topo I inhibitors because a single background cleavage is a built-in control; if the agent stimulates cleavage at this and/or any other sequence in the fragment that contains the hexadecameric site, prominent cutting is readily revealed. Typically, it is difficult to trap topo I cleavages on DNA; one must use more enzyme since cleavage complexes consume the enzyme stoichiometrically. For this reason, higher levels of enzyme are required for analysis of DNA cleavages. The fragment can be isolated from the polylinker using the sites shown in protocols in Maniatis cloning book, end labeled and used as a cleavage substrate.
Relaxed pHOT-1 DNA is shipped at ambient temperature and should be stored a 4°C.
Relaxed pHOT-1 Data Sheet
Relaxed pHOT-1 DNA References
Bonven et al., Cell 41:541 (1985)