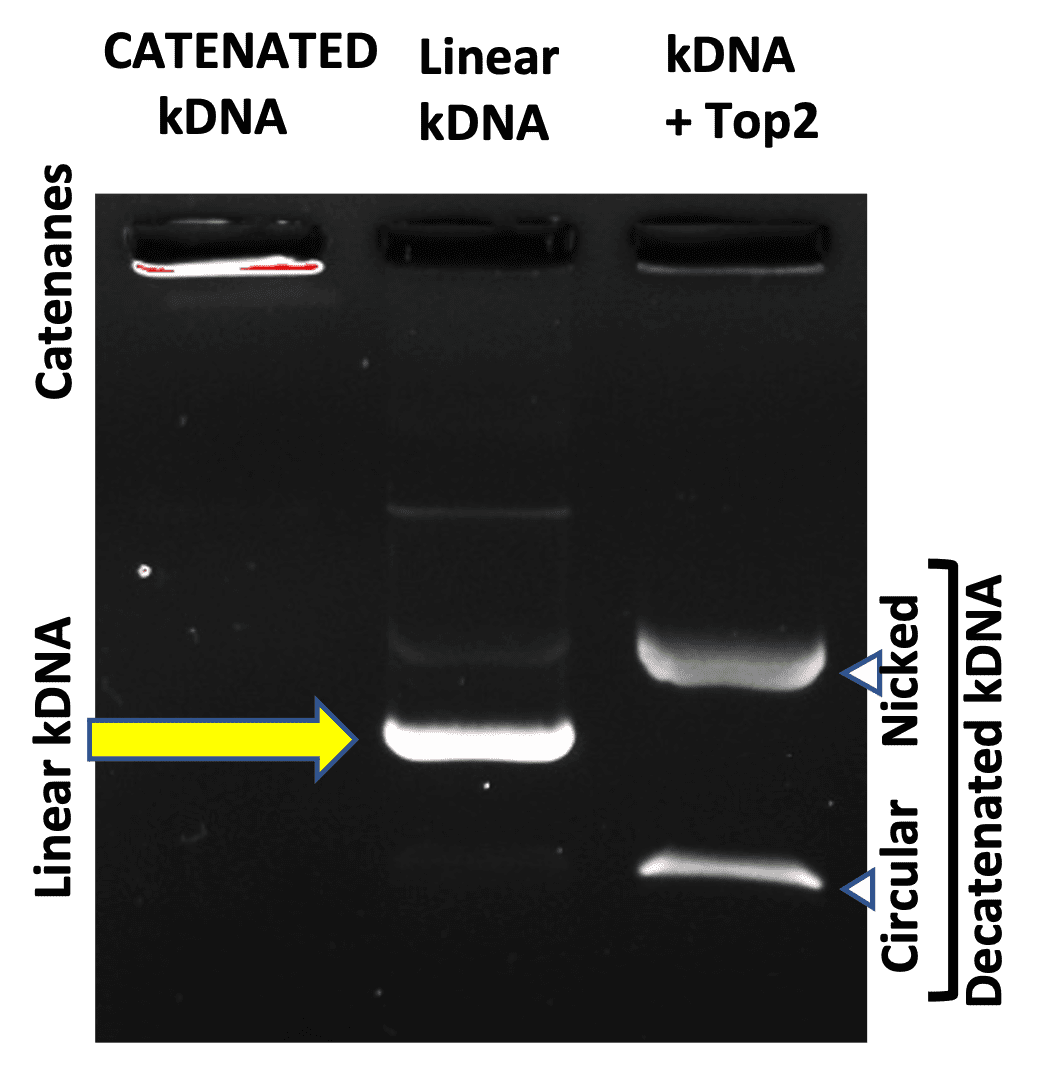
Assays employing crude extracts for topo II activity based upon relaxation of supercoiled DNA can be complicated due to the presence of topo I in partially purified fractions. Additional complications arise with contaminating nuclease activity (due to Mg++) which degrade or nick the supercoiled substrate. These problems can be avoided by using a catenated DNA substrate prepared from the kinetoplast of the insect trypanosome Crithidia fasciculata. KDNA is an aggregate of interlocked DNA minicircles (mostly 2.5 kb) that form extremely large networks of high molecular weight. As a result, these networks fail to enter an agarose gel. Upon incubation with topo II, which engages DNA in a double stranded breaking and reunion cycle, minicircular DNAs are effectively released (decatenated). The decatenated minicircles move rapidly into the gel owing to their small size. This reaction will not occur with topoisomerase I. TopoGEN has modified the original gel system described by Miller et al. to allow additional discrimination of activities using kDNA.
kDNA is an ideal substrate for type II enzymes, both pro- and eukaryote. It is highly specific for topoisomerases that break. both strands and reseal both strands of DNA; this defines a type II (DS DNA) mechanism of action. It is vital that kDNA is clean, purified to a known concentration, free of nucleases and validated to work with top2 enzymes.

Linear kDNA Quality Control Tests
- For catenated KDNA substrate, at least 90% of the DNA will be retained in the well of a 1% agarose gel.
- Decatenation of each batch of kDNA is tested with purified topoisomerase II.
The marker is shipped at ambient temperature and should be stored a 4°C.
References
- Miller et al., J. Biol. Chem. 256:9334-9339 (1981)
- Muller et al., Biochem. 27:8369-8379, 1988
- Muller et al., Nuc. Acid. Res. 17:9499, 1989